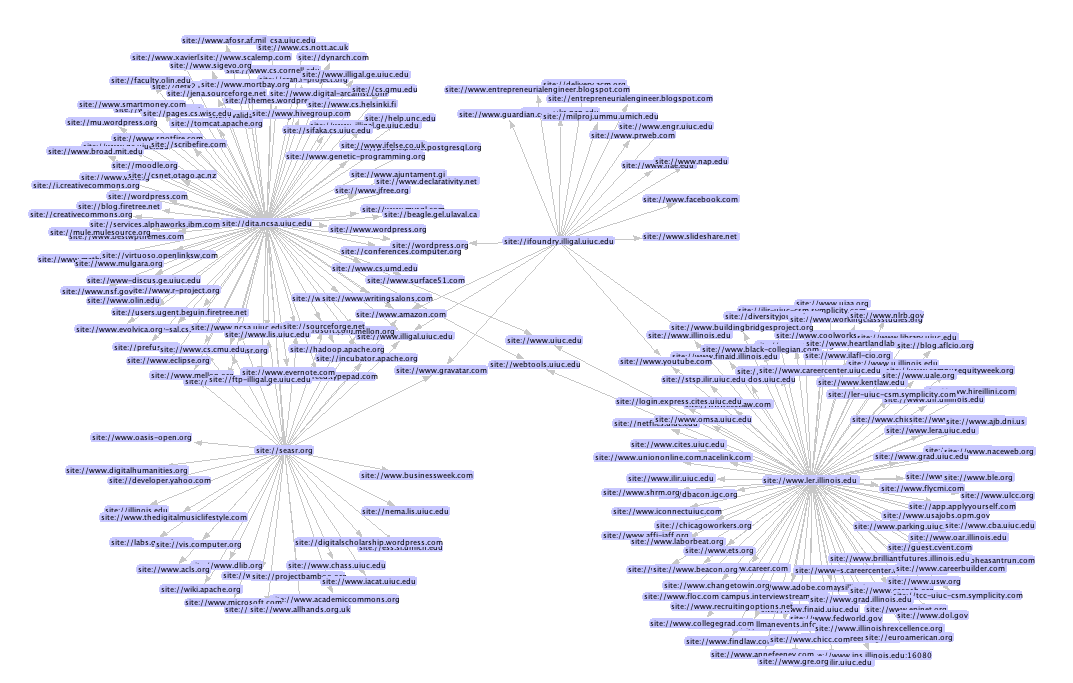
Have you ever wondered who does your intranet link to? I was sitting the other day in a meeting (yes, I know, breaking news) and I was wondering what would be a fast way to be able to answer the question. The basic sketch I did in my mind was simple:
- Set up a web crawler to the domain I want to analyze
- Run the crawling job
- Get the links collected on the web map
- Process the links to only keep the site they refer to
- Remove duplicates
- Visualize the graph
Simple isn’t it? So, what do I need to get it to work? Basically three pieces of software (a web crawler, a graph manipulation library, and a visualization package) and some glue. Going over the things I been playing for the last year I draw three candidates: Nutch, RDFlib, and prefuse. Oh, the glue will be just two Python scripts.
Nutch was the birth place of Hadoop, so I have been running into it but never got the time to play with it. So I did. You can easily restrict the crawl and set a bunch of initial seeds (more information can be found it this tutorial). Once the crawl job finish, nutch provides access to the webmap via the readlinkdb command allowing you to dump the graph of links. And, almost there. The little Python script below parses the dumped file and dumps the site graph into RDF—formated as an N-Triple.
from __future__ import with_statement
def urlToSite ( url ) :
urla = url.split('://')
if len(urla)!=2 :
raise Exception("Unprocessable url ",url)
return urla[1].split('/')[0]
with open("part-00000") as f:
url = None
for line in f:
if line[0]=='\n' :
url = None
elif line[0]!=' ' and line[0:4]!='file' and line[0:6]!='mailto' :
url = urlToSite(line.split("\tInlinks:\n")[0])
elif line[0]==' ' and url!=None :
urlTmp = urlToSite(line.replace(' fromUrl: ','').split(' anchor: ')[0])
print '<site://'+url+'>', '<http://foo.org/incomming>', '<site://+urlTmp+'> .'
Then the RDF can be loaded. RDF statements are unique, thus duplicates are removed. With the graph at hand, it is trivial to serialized it into GraphML (a XML standard for graph serialization) for later visualization. The script below shows this step.
from rdflib.Graph import Graph
g = Graph()
g.parse("part-00000.nt",format="nt")
nodes = {}
for subj in g.subjects() :
nodes[str(subj)]=1
for obj in g.objects() :
nodes[str(obj)]=1
print '<?xml version="1.0" encoding="UTF-8"?>'
print '''<graphml xmlns="http://graphml.graphdrawing.org/xmlns"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://graphml.graphdrawing.org/xmlns
http://graphml.graphdrawing.org/xmlns/1.0/graphml.xsd">'''
print '\t<graph id="G" edgedefault="directed" parse.order="nodesfirst">'
print '\t\t<key id="site" for="node" attr.name="site" attr.type="string">unknown</key>'
for n in nodes:
print '\t\t<node id="'+n+'">'
print '\t\t\t<data key="site">'+n+'</data>'
print '\t\t</node>'
i=0
for stmt in g :
print '\t\t<edge id="e'+str(i)+'" source="'+str(stmt[2])+'" target="'+str(stmt[0])+'" />'
i = i+1
print '\t</graph>'
print '</graphml>'
Finally with the GraphML can be visualized using prefuse—as the screenshot below shows. To run this crawl I used the following seeds http://www.ler.illinois.edu/, http://ifoundry.illigal.uiuc.edu/, http://dita.ncsa.uiuc.edu/, and http://seasr.org/.